Some sequence motifs in proteins tend to be modified. Thus, for example an N-terminal glutamine or glutamic acid residue can spontaneously lead to a cyclization. This reaction results in an N-terminal pyroglutamic acid with elimination of ammonia or water, respectively. Aspartyl and asparaginyl deamidation, isomerization, and racemization reactions can spontaneously occur, if the subsequent amino acid is glycine. This could alter the protein structure and thereby inhibit the antigen binding affinities or lead to a backbone hydrolysis. In eukaryotic cells, NXS/T motifs are predominately glycosylated, where X denotes any amino acid except proline. All alleles and isotypes of human IgG γ-chains terminates with lysine. In most cases these C-terminal lysines were enzymatically processed by basic carboxypeptidases.
Al these modifications are triggered by a specific sequence motif. This allows Prot pi | Protein Tool making predictions of potential modifications sites by means of the amino acid sequence. Figure 1 shows the amino acid sequence of the heavy chain of trastuzumab (a therapeutic monoclonal antibody) in the results section of a calculation request on Prot pi | Protein Tool. With the drop-down menu beneath the sequence, modification sites can be selected to be highlighted. In my last post you can find a detailed tutorial how to enter the amino acid sequence and run a calculation.
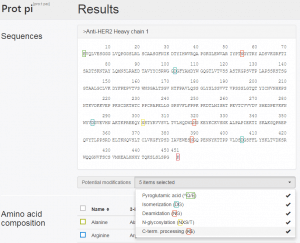
Trastuzumab has been extensively analyzed by Yi Wang (2012) from Northeastern University using LC-MS based approaches. In this respect it was found that the heavy chain of trastuzumab has undergone all these common chemical modifications.
Table 1: Predicted and measured modifications of the heavy chain of trastuzumab with the percentage of appearence.| Modification | Seq. Loc. | Predicted | Measured | Percentage |
|---|---|---|---|---|
| Pyroglutamate | 1 (E) | ✔ | ✔ | 1.32 % |
| Deamidation | 55 (N) | ✔ | ✔ | 2.93 % |
| Deamidation | 84 (N) | ✘ | ✔ | 0.19 % |
| Deamidation | 319 (N) | ✔ | ✔ | 1.51 % |
| Deamidation | 388 (N) | ✔ | ✔ | 2.22 % |
| Isomerization | 102 (D) | ✔ | ✔ | 0.61 % |
| Isomerization | 284 (D) | ✔ | ✘ | n.a. |
| Isomerization | 405 (D) | ✔ | ✔ | 0.92 % |
| N-Glycosylation | 301 (N) | ✔ | ✔ | 100 % |
The deamidation of asparagine 84 was the only modification which was measured but not predicted. However, the degree of deamidation at this site was only very small 0.19 %. The other way around, the aspartic acid 284 was identified as potential modification site, but no modification was observed. Nevertheless the predicted and the measured modification sites correlate very well. Thus, this is a straightforward feature to find potential modification sites, and hence to focus on the relevant further investigations in the laboratory.
Thank you for your trust in Prot pi!
[ratings]