ADP-ribosylation is a reversible post-translational modification in which one (mono-ADP-ribosylation) or multiple (poly-ADP-ribosylation) ADP-ribose moieties are attached onto a substrate protein by ADP ribosyltransferases.
mono-ADP-Ribosylation
Overview
mono-ADP-ribosylation is a common post-translational modification, where an ADP-ribose moiety is transferred from NAD+ to the substrate protein under the release of nicotinamide. The transfer of ADP-ribose occurs onto amino acid residues with a nucleophilic oxygen, nitrogen or sulfur.
| pKa | NC | Loss | Gain | Deltamass | H | AA | UV-Spec | Pattern |
| – | No | H | C15H22 N15O13P2 | Av: 541.3011 M: 541.0611 | – | C, D, E, K, N, Q, R, S, T | Yes* | – |
*Spectrum of ATP instead of ADP-ribose was taken since no spectrum of ADP-ribose was available. The major difference between the two molecules are the terminal phosphate/ribose group. The absorbance in the UV-range however is mainly caused by the adenine moiety.
In-depth mechanism
mono-ADP-ribosylation describes the process of attaching an ADP-ribose moiety directly onto an amino acid residue with nucleophilic oxygen, nitrogen or sulfur, forming an N-, O- or S-glycosidic bond. The donor of the ADP-moiety is nicotinamide adenine dinucleotide. The transfer is catalysed by various ADP-ribosyltransferasen. During the transfer, nicotinamide is released1–6. Primarily, arginine 1–6, glutamate1–5 and aspartate1–5, but to a lesser extent also asparagine1,2,5,6, lysine1–4, cysteine1,3,6, threonine3, glutamine6 and histidine6 residues have been described as acceptor for ADP-ribosylation. ADP-ribosylation increases the molecular mass of the modified protein by 541 Da2. Furthermore, an additional negative charge is introduced into the protein1. The mechanism of mono-ADP-ribosylation is shown in figure 1.
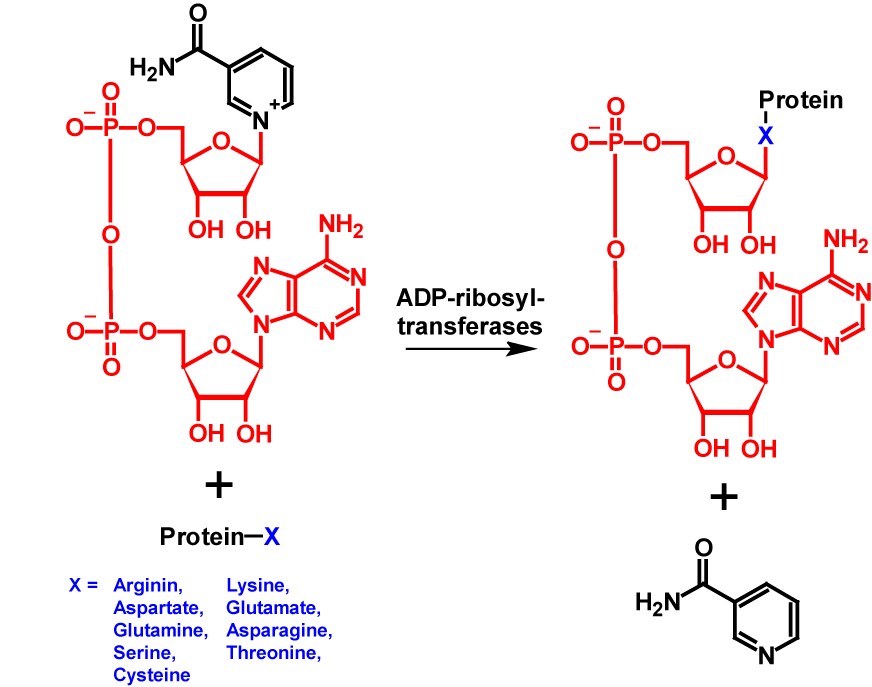
It is assumed that mono-ADP-ribosylation was originally developed by bacteria as a defence mechanism against viruses, other bacterial species and antimicrobial molecules3. Otherwise not much is known about the functions of mono-ADP-ribosylation2.
poly-ADP-ribosylation
Some ADP-ribosyltransferases are able to perform several ADP-ribosylations, which is called poly-ADP-ribosylation. In poly-ADP-ribosylation, further ADP-ribose moieties are added to the already protein-bound ADP-ribose, thus forming ADP-ribose polymers4,7. Chain elongation occurs via a nucleophilic attack of the C2″ hydroxyl of the adenine ribose on the C1″ of the next nicotinamide adenine dinucleotide4. The polymers may be irregularly branched, with a branching point every 20 – 60 units of linear ADP-ribose7. Since each ADP-ribose moiety adds a negative charge to the protein, a protein that previously was positively charged may no longer be charged or may even become negatively charged1. In contrast to mono-ADP-ribosylation, poly-ADP-ribosylation plays a crucial role in various major cellular and biological processes, such as DNA damage repair, cell proliferation and differentiation, metabolism, stress and immune responses2,5.
References
- 1.Simonet NG, Rasti G, Vaquero A. The Histone Code and Disease. In: Epigenetic Biomarkers and Diagnostics. Elsevier; 2016:417-445. doi:10.1016/b978-0-12-801899-6.00021-8
- 2.Martello R, Leutert M, Jungmichel S, et al. Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat Commun. Published online September 30, 2016. doi:10.1038/ncomms12917
- 3.Cohen MS, Chang P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat Chem Biol. Published online February 14, 2018:236-243. doi:10.1038/nchembio.2568
- 4.Lüscher B, Bütepage M, Eckei L, Krieg S, Verheugd P, Shilton BH. ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease. Chem Rev. Published online November 27, 2017:1092-1136. doi:10.1021/acs.chemrev.7b00122
- 5.Palazzo L, Mikolčević P, Mikoč A, Ahel I. ADP-ribosylation signalling and human disease. Open Biol. Published online April 2019:190041. doi:10.1098/rsob.190041
- 6.Green KD, Garneau-Tsodikova S. Posttranslational Modification of Proteins. In: Comprehensive Natural Products II. Elsevier; 2010:433-468. doi:10.1016/b978-008045382-8.00662-6
- 7.Hassa P O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. Published online 2008:3046. doi:10.2741/2909
Hallo Jon,
schöne Abb. Setze ich für meine Vorlesung ein. Ergänzung: Diphtherietoxin ADP-ribosyliert das His715 im EF-2.
VG
Andreas Rummel